Confirmation of computational grids for patient-specific biomechanical models to predict the brain deformations for image-guided neurosurgery
Contents
Background
The importance of intra-operative image registration (computational radiology) is growing rapidly as new therapeutic technologies that are entering the medical practice now and will be employed in the future, such as nanotechnological devices, drug polymers, robotic surgery and robotic prosthetics, all have extremely localised areas of therapeutic effect. As a result, they have to be applied precisely in relation to the patient’s current (i.e. intra-operative) anatomy, directly over the specific location of anatomic or functional abnormality.
As only pre-operative anatomy of the patient is known precisely from medical images (usually MRI), it is now recognised that the ability to predict soft organ deformation (and therefore intra-operative anatomy) during the operation is the main problem in performing reliable surgery on this.
The ISML is particularly interested in problems arising in image-guided neurosurgery. In this context it is very important to be able to predict the effect of procedures on the position of pathologies and critical healthy areas in the brain. If displacements within the brain can be computed during the operation, they can be used to warp high-quality pre-operative MR images so that they represent the current configuration of the brain.
Introduction
We have a large collaborative project with Harvard that aims at demonstrating that our biomechanics-based methods generate information that aids neuronavigation. Within the project 50 cases of neurosurgery need to be analysed. Each case is documented by the preoperative MRI, intraoperative 1 MRI taken just after craniotomy (but before anything has been done to the brain), intraoperative 2 MRI taken for some cases during the procedure, and postoperative MRI.
Pipeline
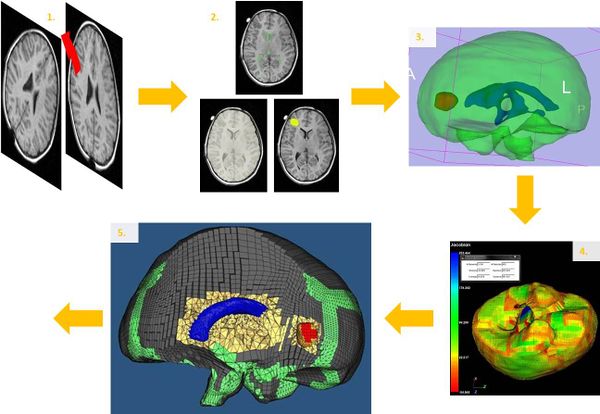
The following steps have to be done on each case:
- Resampling and registration of the preoperative and intraoperative-1 images in order to bring the two images into a common coordinate frame.
- Segmentation of the brain, ventricles and the tumour by using 3D-Slicer ( https://www.slicer.org ).
- Modelling of the brain, ventricles and the tumour in 3D.
- Meshing the model with blocks by using IA-FEMesh (IA-FEMesh Center for Computer Aided Design).
- Improvement of the meshing and insertion of ventricles and the tumour inside the brain by using HyperMesh (Software include in HyperWorks from Altair).
- Definition of the loading with an accurate segmentation of the exposed brain surface (due to the craniotomy) on the intraoperative-1 MRI.
- Computation of the deformation by using the algorithm written by Research Assistant Prof. Grand Joldes.
- Evaluation of results
Resources
File:Presentation of Jeremie VOLTZ (5th July 2011).pdf